Help
Curated Database
GlyConnect is a curated database, collecting information in publications, from which pieces of evidence that describe specific aspects of glycosylation are extracted. Here's how we build it:
- Starting with Publications: We select and review scientific publications that focus on glycosylation and related topics.
-
Extracting Evidence: From these publications, we identify pieces of evidence that describe specific aspects of glycosylation.
Each piece of evidence are related to one or more of these:
- A glycan with its structural details, when available.
- The glycosylation of a protein.
- Glycosylation events in specific tissues, cell types, diseases, or organisms.
-
Knowledge Graph: This information is organised as a Knowledge Graph, where:
- Each Publication is linked to multiple Evidence nodes.
- Each Evidence node is further connected to other entities, such as :
- Protein the glycosylated protein - more specifically a glycoprotein
- Glycan Structure / Monosaccharide the structural details of a glycan
- Taxon / Tissue / Cell Type / Cell Component / Cell Line the biological source(s) in which glycosylation was characterised
- Disease the disease in which glycosylation was characterised
The list of nodes connected to Evidence nodes are listed below with their colour codes:
- Monosaccharide
- Glycan structure
- Protein
- Taxon
- Disease
- Tissue
- Cell component
- Cell type
- Cell line
- Publication
This structure allows us to organize complex scientific information into clear, searchable relationships, helping you find precise answers to your queries.
GlyConnect is developed using the Memgraph graph database management system, taking advantage of its capabilities for efficient storage and querying of graph-structured data. The Database Schema is visible upon clicking on the upper right of the search homepage.
Search Engine
The graph database search system is built on Cypher®, the graph query language developed for Neo4j. The engine may run in three modes:
- the full-text search performing search in entire text of documents across various data types,
- the guided search suggesting queries in a path-based syntax,
- the advanced search processing Cypher queries for more complex operations.
Full text search
Enter terms to enable the search engine to conduct a full-text search across one or more node types.
Please refer to the Pattern Matching System section below for more details.
One or more data types must be selected for searching a term. The invert button can be used to quickly select multiple types at once.
Only the graph nodes matching the search can be presented in the results.
For example:
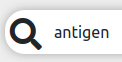 searches any data matching the term
searches any data matching the term antigen. searches everything across all data types.
searches everything across all data types.
 searches for protein data that match the term
searches for protein data that match the term antigen.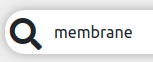
 combined searches for tissues, cell components, cell types and cell lines data that match the term
combined searches for tissues, cell components, cell types and cell lines data that match the term
membrane.
Guided search
The graph query language includes an intuitive builder that streamlines the query creation process. In guided search mode, both graph nodes and edges are searchable. As keywords are typed, the system offers suggestions for relevant node types, field names, accessible nodes, and even stored Cypher queries. At each step, the selected term must be validated to progressively build the query.
Please refer to the Pattern Matching System section below for more details.
For example:
- (Taxon) search all nodes of type
Taxon. - (Taxon:human) search all nodes of type
Taxonthat contain the term 'human' - (Taxon.name:human) search all nodes of type
Taxonwhere the fieldnamecontains the term 'human' - (Taxon.name:human)--(CellComponent) search all paths from 'human'
Taxonnodes to anyCellComponentnodes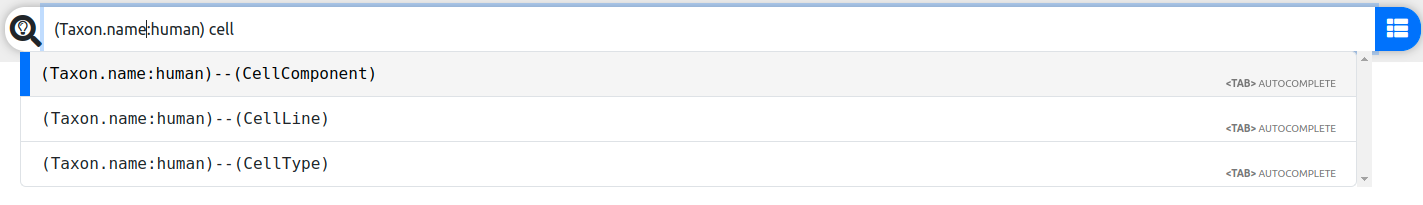
Advanced search
Cypher® is Neo4j’s declarative query language, allowing users to unlock the full potential of property graph databases. We have chosen Memgraph as our graph database solution which allows for optimized performance and faster query execution, especially for real-time data handling. Memgraph uses openCypher an open source implementation of Cypher® but we will refer to it as Cypher for simplicity.
The advanced search mode relies on graph query methods for optimal searching. The documentation on Cypher syntax can be found here.
A selection of stored queries can be accessed by clicking from the sidebar area.
Pattern Matching System (available only for Basic and Guided search)
The search query system allows for flexible searches using special symbols to define term relationships.
Disjunctive (OR) Search
A space between terms indicates a disjunctive search, where at least one term must appear in the document.
For example:
- immunoglobulin gamma will match documents containing either
immunoglobulin,gamma, or both.
Conjunctive (AND) Search
The + symbol defines a conjunctive search, requiring all terms to be present.
For example:
- immunoglobulin+gamma will match documents containing both
immunoglobulinandgamma.
Anchors (available only for Guided field search)
Anchors refine searches by specifying term positions:
- The
>anchor ensures the term appears at the beginning of the document. - The
<anchor forces the term to appear at the end of the document.
For example:
- (Protein.name:>immunoglobulin<) will match documents that is exactly
immunoglobulin. - (Protein.name:>immunoglobulin) will match documents that starts with
immunoglobulin. - (Protein.name:immunoglobulin<) will match documents that ends with
immunoglobulin. - (Protein.name:immunoglobulin) will match documents where
immunoglobulinappears anywhere in the text.
Search Results navigation
We currently support three navigation modes: paginated view, infinite scrolling, and graph view.
- Paginated View Displays data in separate pages with navigation links for easy browsing
- Infinite Scrolling Automatically loads more content upon scrolling down, providing a continuous browsing experience
- Graph View Visualizes data in a graph format to emphasise relationships and patterns interactively
Guided Tours
For a more immersive experience, we offer several guided tours, three of which explain the three search modes. These tours are accessible by clicking the directions icon at the top left. If any tours are incomplete, the icon will blink and indicate how many tours are still available.