Example queries using curl
Write a command including the query to select 10 predicates and return data as json :
curl -X POST https://glyconnect.expasy.org/glystreem/sparql --data-urlencode 'query=SELECT * { ?s ?p ?o
} LIMIT 10' --data-urlencode 'format=json'
The SPARQL query can be loaded from a file (count_structures_tn_antigen.rq in the example):
curl -X POST https://glyconnect.expasy.org/glystreem/sparql -H 'content-type: application/sparql-query'
-H 'accept: application/sparql-results+json' --data-binary @count_structures_tn_antigen.rq
Output formats
- application/sparql-results+json
- application/sparql-results+xml
- text/csv
- text/tab-separated-values
Sample SPARQL files
- quant_1_count_structures_substituent_root.rq
- Count of all structures that only have a substituent in the root node
- quant_2_count_glycans_residueroot.rq
- Count of all glycans that have a ResidueRoot
- quant_3_count_unique_basetypes_residueroot.rq
- Count of all unique base types found in ResidueRoot
- quant_4_count_unique_basetypes.rq
- Count of all unique base types
- quant_5_count_unique_substituents.rq
- Count of all unique substituent types
- quant_6_count_unique_substituents_single_residue.rq
- Count of all unique substituent types that are a single residue (not part of a composed residue)
- quant_7_count_structures_tn_antigen.rq
- Count of all di-sialyl Tn antigen type structures
- quant_8_count_structures_fuc_root.rq
- Count of all structures that start with Fucose
- quant_9_count_structures_man_root.rq
- Count of all structures that start with Mannose
- quant_10_count_structures_xyl_root.rq
- Count of all structures that start with Xylose
- quant_11_count_structures_bagitem_atleast.rq
- Count of all structures that have from (at least) 1 to 9 undefined sections (bag items)
- quant_12_count_structures_bagitem_equals.rq
- Count of all structures that have exactly 1 to 9 undefined sections (bag items)
- quant_13_count_structures_no_glycancore.rq
- Count of all structures that have no associated GlycoCT string (therefore no GlycanCore)
- quant_14_count_structures_glycan.rq
- Count of all structures that have associated Glycan
- quant_15_count_structures_glycancore.rq
- Count of all structures that have associated GlycanCore
- quant_16_count_structures_residue_root.rq
- Count of all structures that have associated ResidueRoot
- qual_1_structures_nlinked_glcnac2man3_core.rq
- Display IDs for structures matching exactly N-linked GlcNAc2Man3 core
- qual_2_structures_with_glcnac_phosphate.rq
- Display IDs for structures with exactly GlcNAC and phosphate
- qual_3_structure_671_bagitems.rq
- Show the undefined sections (bag items) for structure 671
- qual_4_linked_sialic_acid.rq
- Does the database contain any structures with two (or more) sialyl acids consecutively linked?
- qual_5_olinked_core1_chained_sialic_acid.rq
- Does the database contain any O-linked Core 1 with two sialyl acids consecutively linked?
- qual_6_olinked_monosaccharides.rq
- Pull out only O-linked monosaccharides
Federated Queries & GlycoQL Query Patterns
- Query 1.1
- List all GlyConnect protein ids, site, no. of glycans, & list of glycans for COVID from GlyConnect & GlySTreeM
- Query_1.2
- Pull out site, and glycosylation related comments for UniProt accession no. P0DTC2 from UniProt
- Q2_1-GlycoQL Hybrid Pattern
- GlycoQL generated pattern to query for N-linked hybrid type structures
- Q2.2-GlycoQL High Mannose Pattern
- GlycoQL generated pattern to query for N-linked high mannose type structures
- Q2.3-GlycoQL GlcNAc Pattern
- GlycoQL generated pattern to query for N-linked mono-GlcNAc type structures
- Q2.4-GlycoQL Complex, Tetra-antennary, Sialylated Pattern
- GlycoQL generated pattern to query for N-linked complex, tetra-antennary, sialylated type structures
- Q2.5-GlycoQL Complex, Tetra-antennary, Neutral Pattern
- GlycoQL generated pattern to query for N-linked complex, tetra-antennary, neutral type structures
- Q2.6-GlycoQL Complex, Tri-antennary, Sialylated Pattern
- GlycoQL generated pattern to query for N-linked complex, tri-antennary, sialylated type structures
- Q2.7-GlycoQL Complex, Tri-antennary, Neutral Pattern
- GlycoQL generated pattern to query for N-linked complex, tri-antennary, neutral type structures
- Q2.8-GlycoQL Complex, Bi-antennary, Neutral Pattern
- GlycoQL generated pattern to query for N-linked complex, bi-antennary, neutral type structures
- Q2.9-GlycoQL Complex, Bi-antennary, Sialylated Pattern
- GlycoQL generated pattern to query for N-linked complex, bi-antennary, sialylated type structures
DataSets
Performance Times
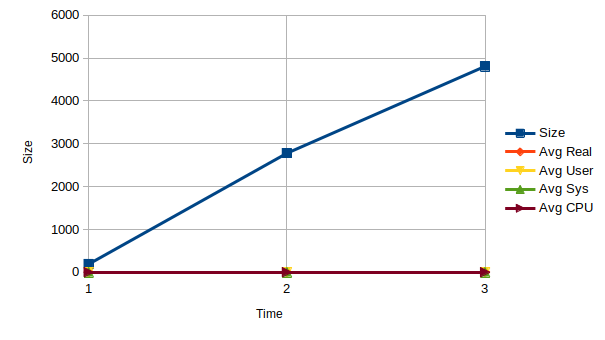
To estimate the running time of SPARQL queries we tested against three triple stores of different sizes, 191, 2790 & 4808 structures. Real time is wall clock time, and user and system times together give total CPU time. These runs were timed on a DELL Latitude 7140 running Ubuntu 20, GraphDB 9.9.0 and python 3.9 (to automize the queries). There were 33 queries run, of which a subset are presented in the paper (available on this page). As can be seen from the table, the CPU time varies little between the different size datasets. The real time increases linearly with the increase in size.
Using extrapolation we predict a real time of just under 1.5 minutes to run 30 queries on a dataset of 100,000 structures (see chart). Data point 1 = Dataset 1 (a, b), Data point 2 = GlySTreeM (c, d), extrapolate to x (100,000 structures) using formula,
`f(x) = b + (x - a) * (d - b)/(c - a)`
| Size | Avg Real Time (s) | Avg User Time (s) | Avg Sys Time (s) | Avg CPU Time (s) | |
|---|---|---|---|---|---|
| Dataset 1 | 191 | 0.71 | 0.272 | 0.025 | 0.297 |
| Dataset 2 | 2780 | 2.713 | 0.259 | 0.029 | 0.288 |
| GlySTreeM | 4808 | 4.544 | 0.262 | 0.017 | 0.279 |
| Extrapolate | 100,000 | 83.592 | 0.056 | -0.148 | -0.092 |
The 30 queries were also run on a remote instance of GlySTreeM - this took 9.5 seconds in real time and 0.5 seconds total CPU time. Using the above table we estimate that it would take just under 3 minutes to run the queries on a remote instance of a 100000 size dataset. However, this needs to be confirmed.